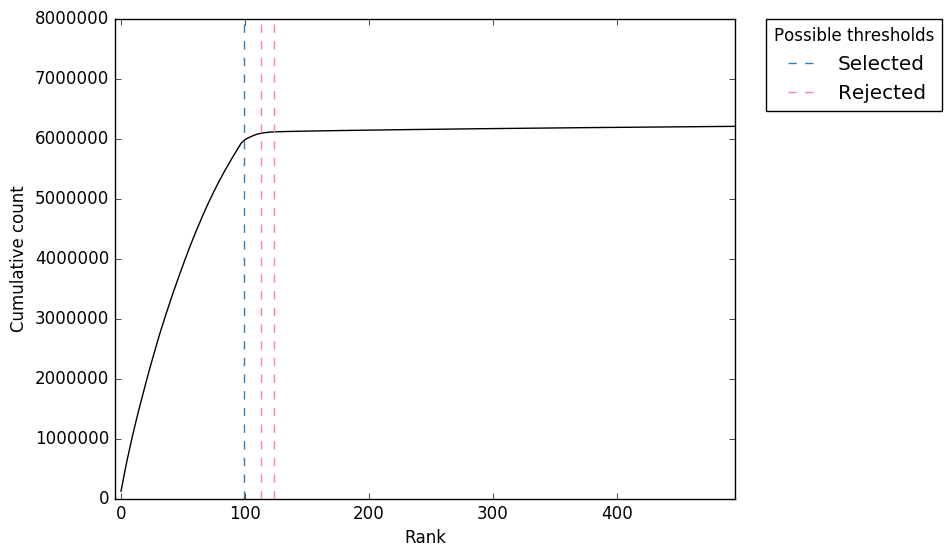
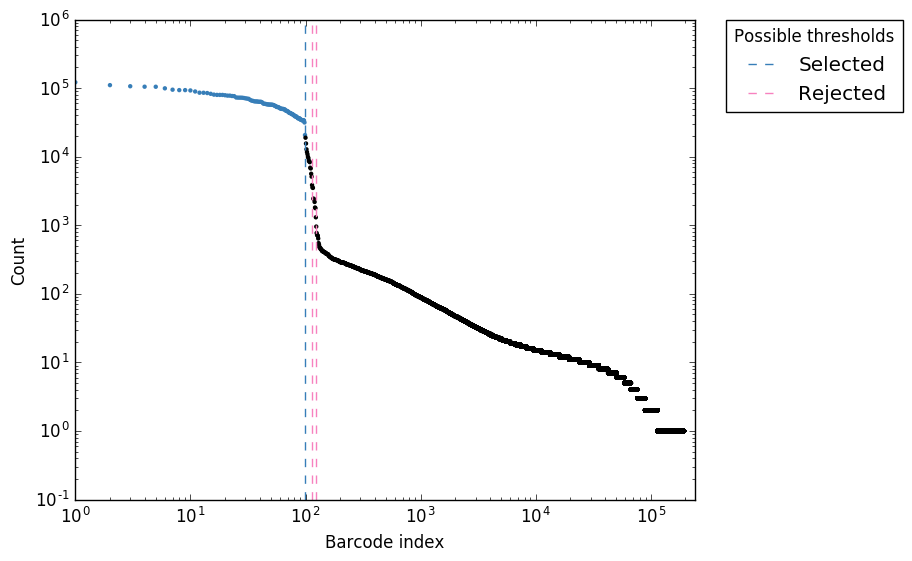
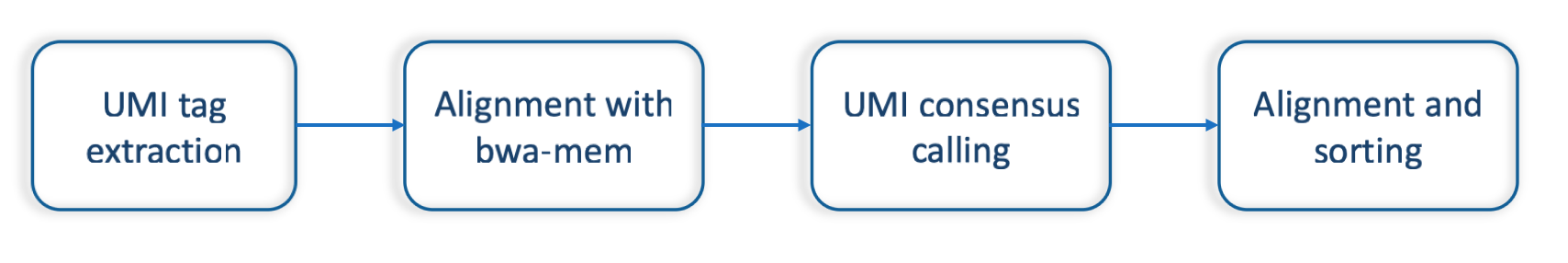
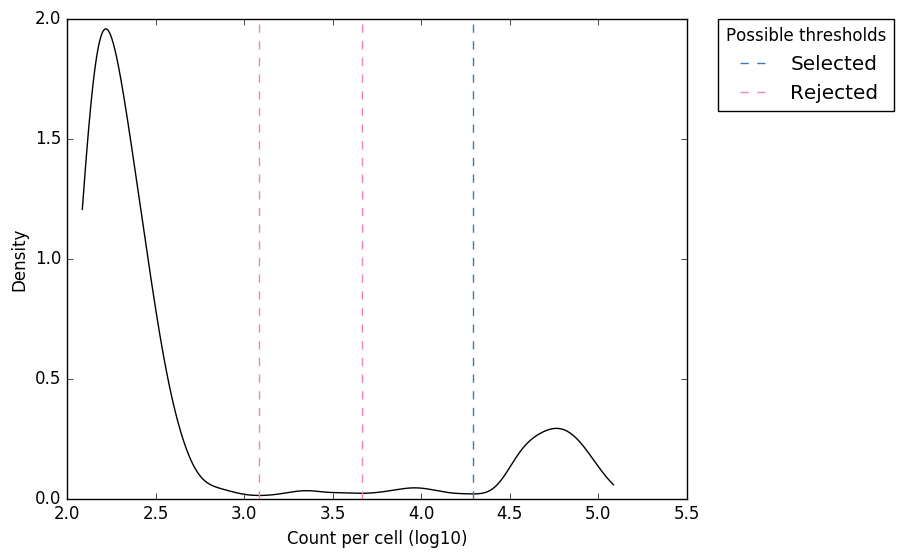
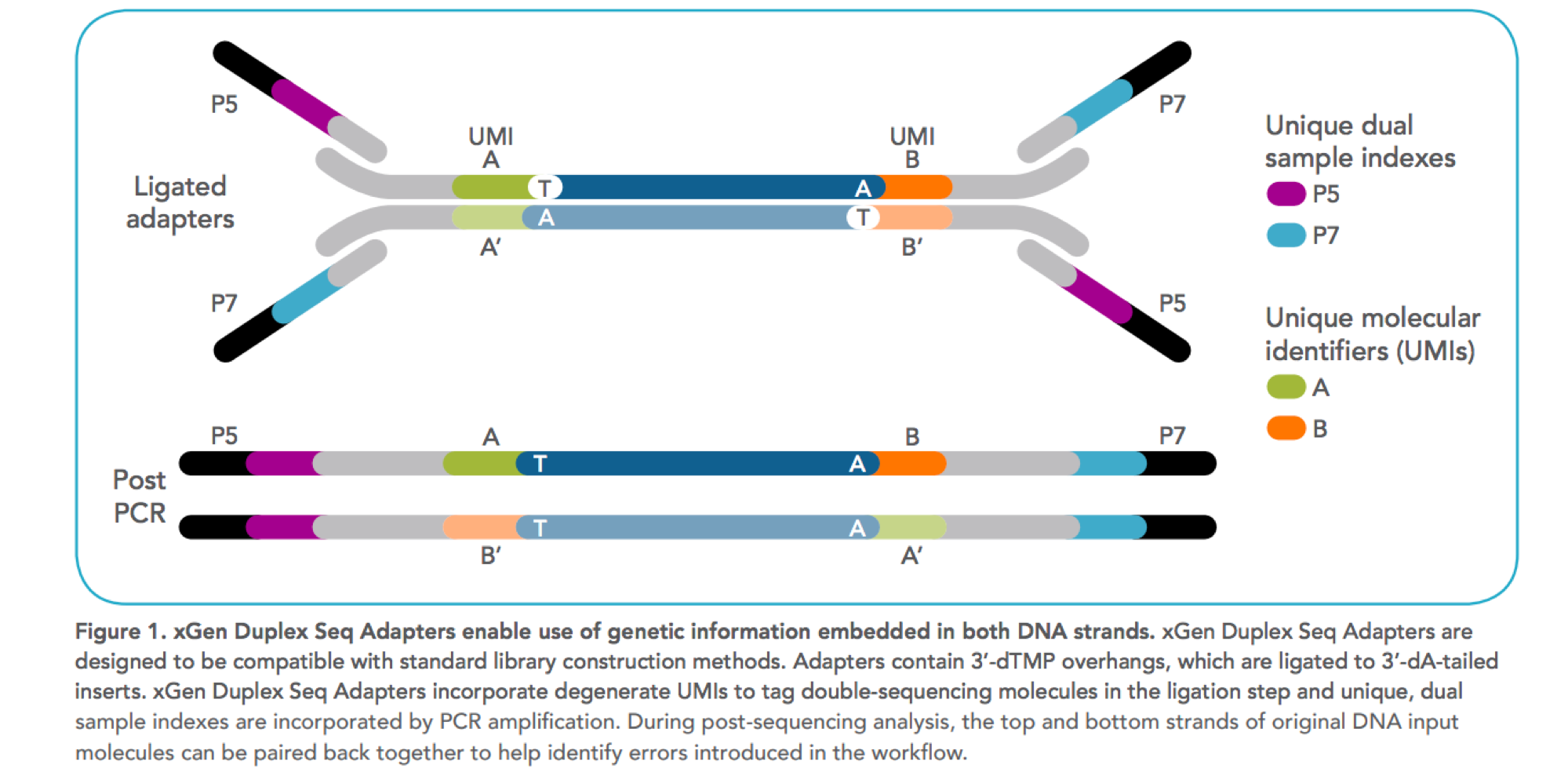
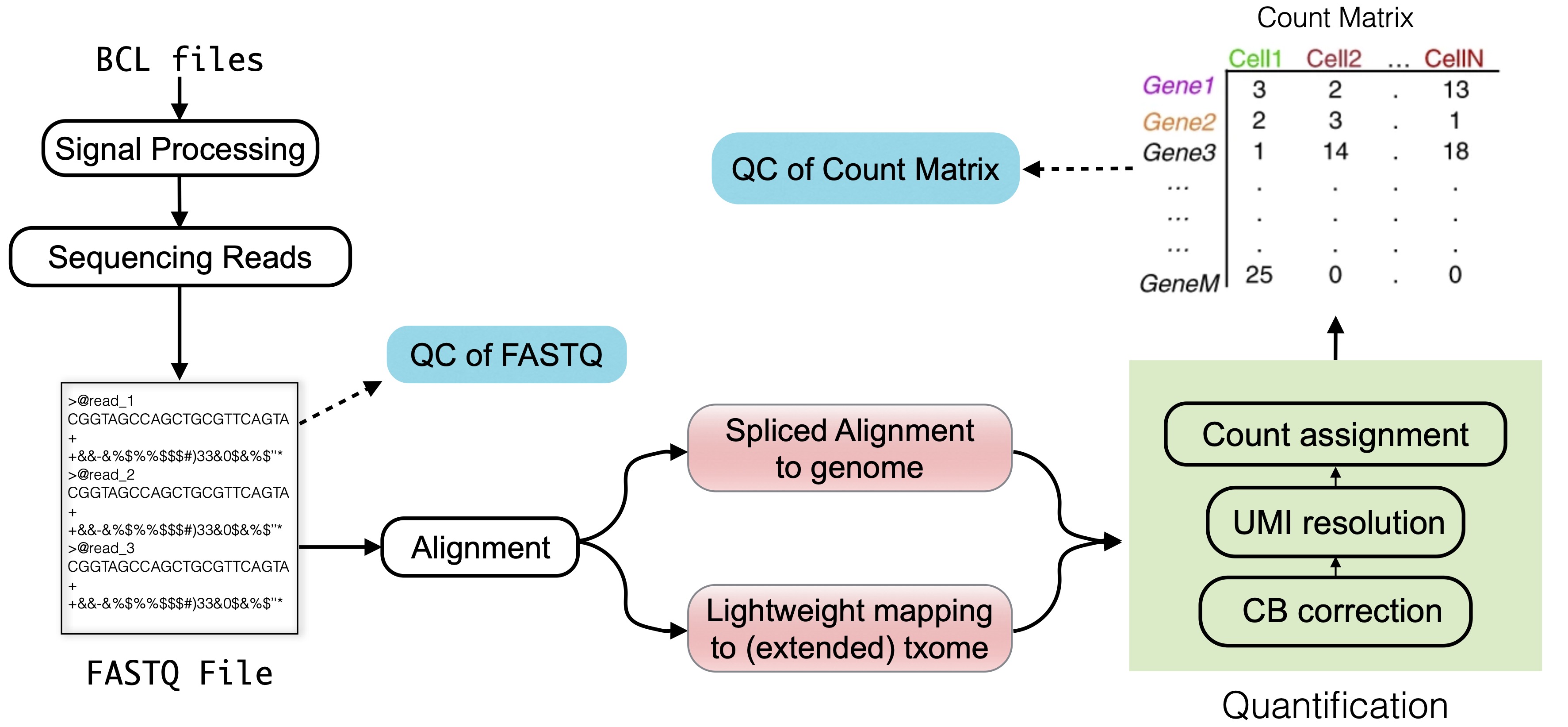
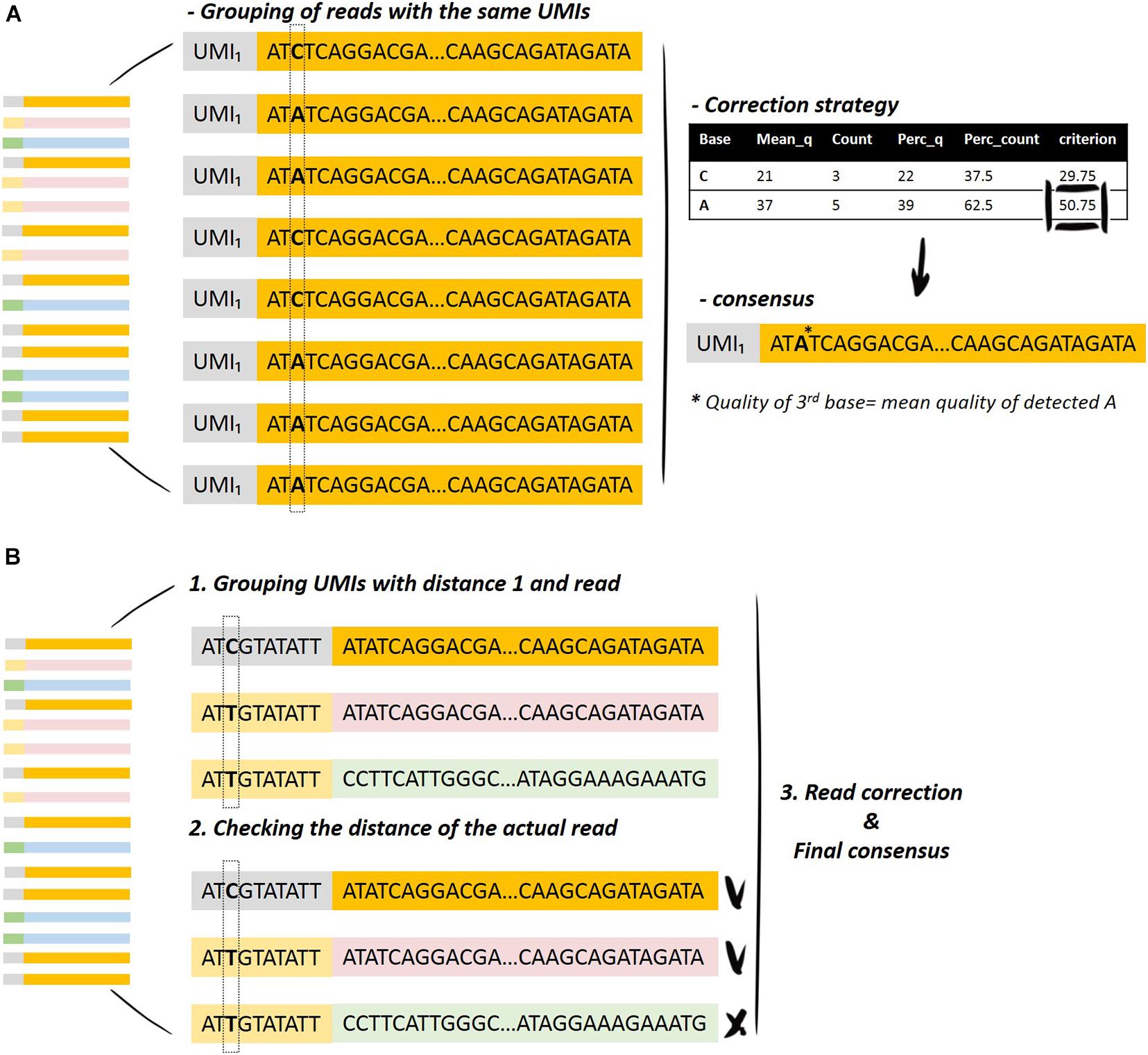
Software input: UMI-VarCal can handle raw and UMI-extracted BAM or SAM | Download Scientific Diagram

Comparison of speed, memory peak and processing results of different... | Download Scientific Diagram

UMI-tools: Modelling sequencing errors in Unique Molecular Identifiers to improve quantification accuracy | bioRxiv
how to extract UMI barcode from .FASTQ file(GSM3139597)? · Issue #356 · CGATOxford/UMI-tools · GitHub
UMI-tools: modeling sequencing errors in Unique Molecular Identifiers to improve quantification accuracy